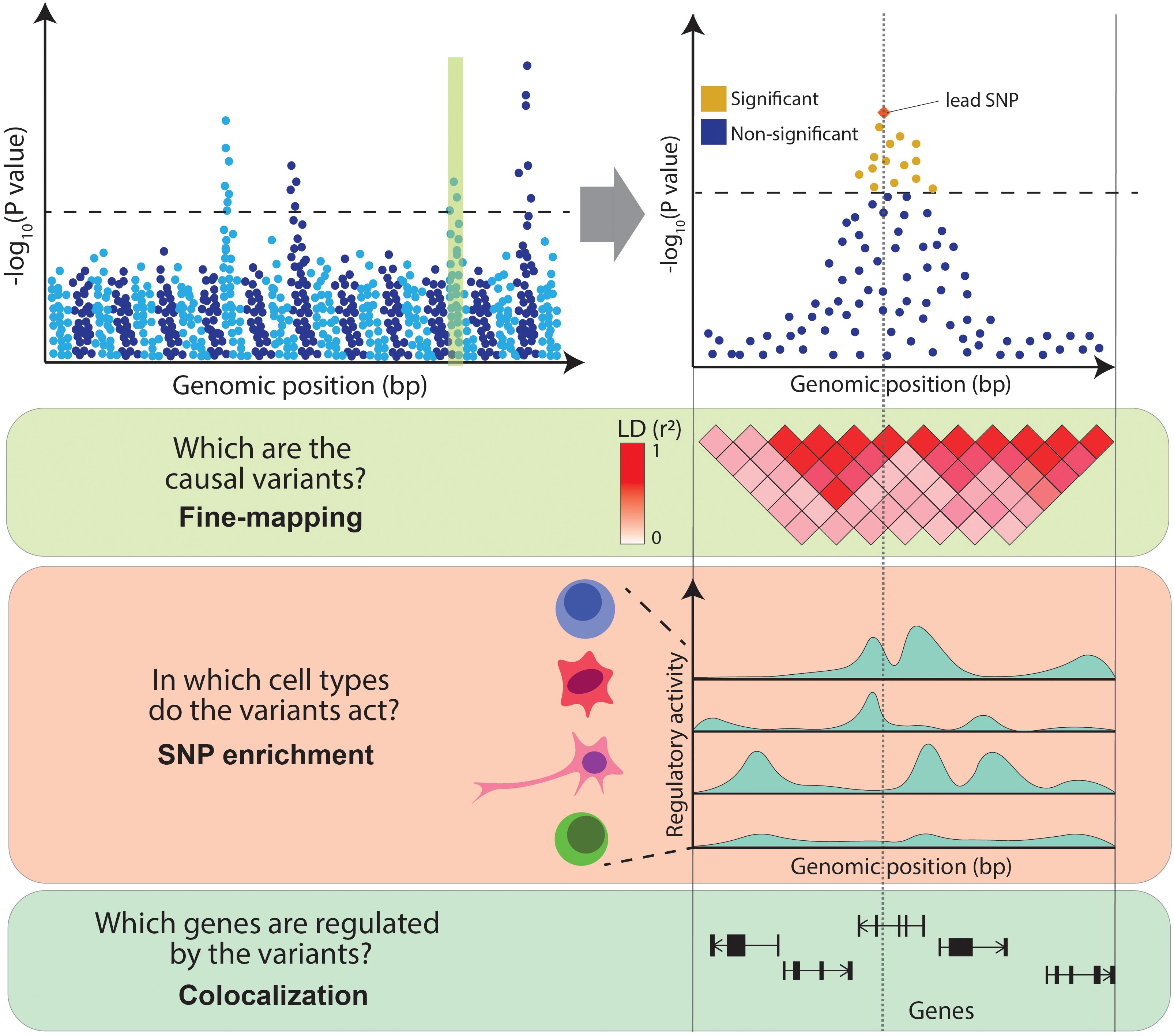
Fine Mapping Gwas
Fine Mapping Gwas – We outline a general coalescent framework for using genotype data in linkage disequilibrium-based mapping studies. Our approach unifies two main goals of gene mapping that have generally been treated . Suggestive signals were followed-up for meta-analysis with a previous GWAS (2751 cases/15 886 controls). We tested for enrichment of association signals in a broad range of functional annotations, and .
Fine Mapping Gwas
Source : www.nature.com
MCB 182 Lecture 12.9 Fine mapping causal variants based on GWAS
Source : m.youtube.com
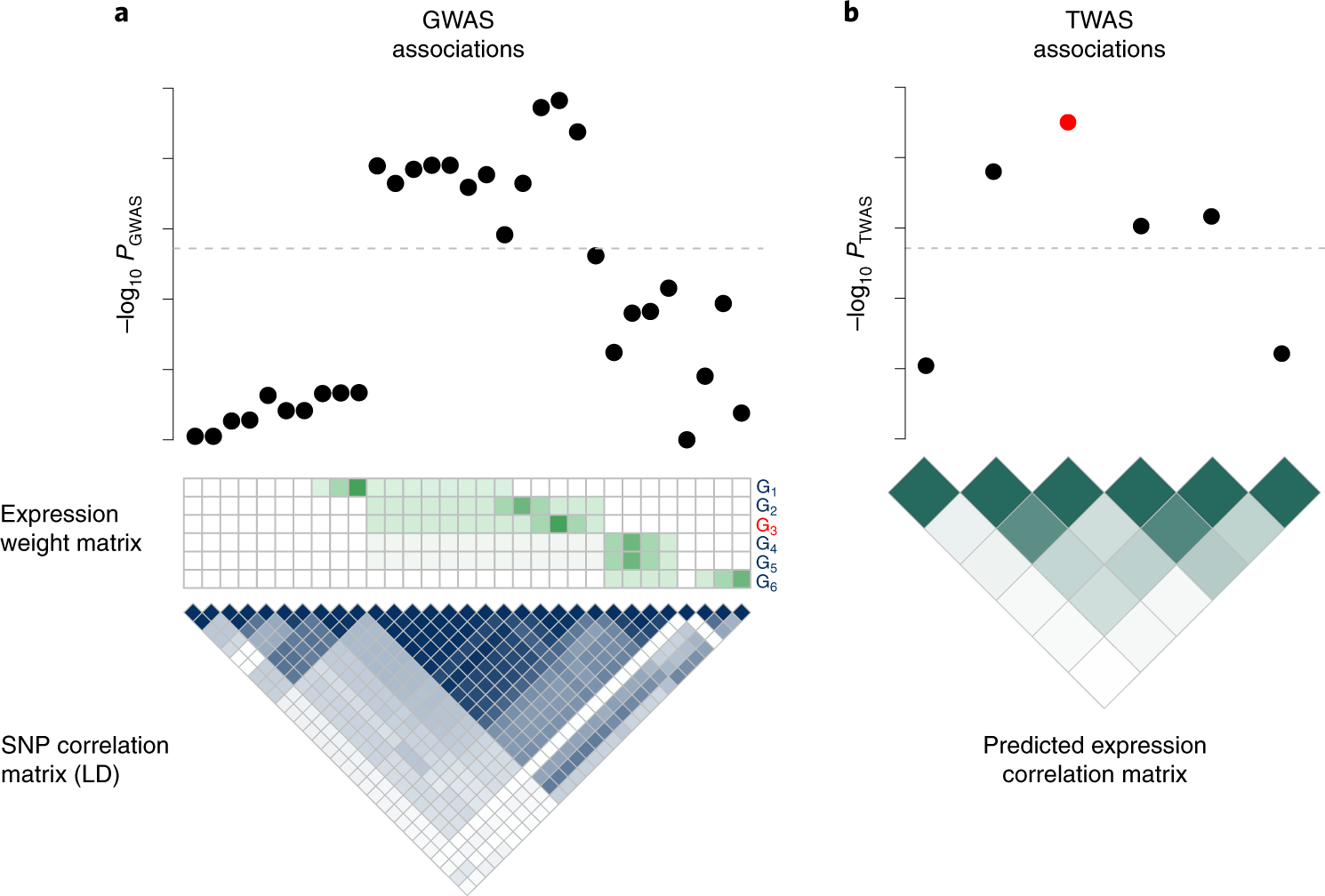
Probabilistic fine mapping of transcriptome wide association
Source : www.nature.com
Fine mapping Basics GWASTutorial
Source : cloufield.github.io
Methods for statistical fine mapping and their applications to
Source : link.springer.com
Fine mapping GWAS loci of gestational duration. (A) PIPs of SNPs
Source : www.researchgate.net
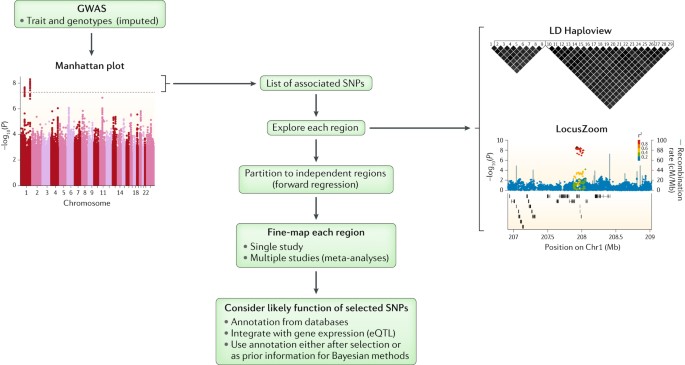
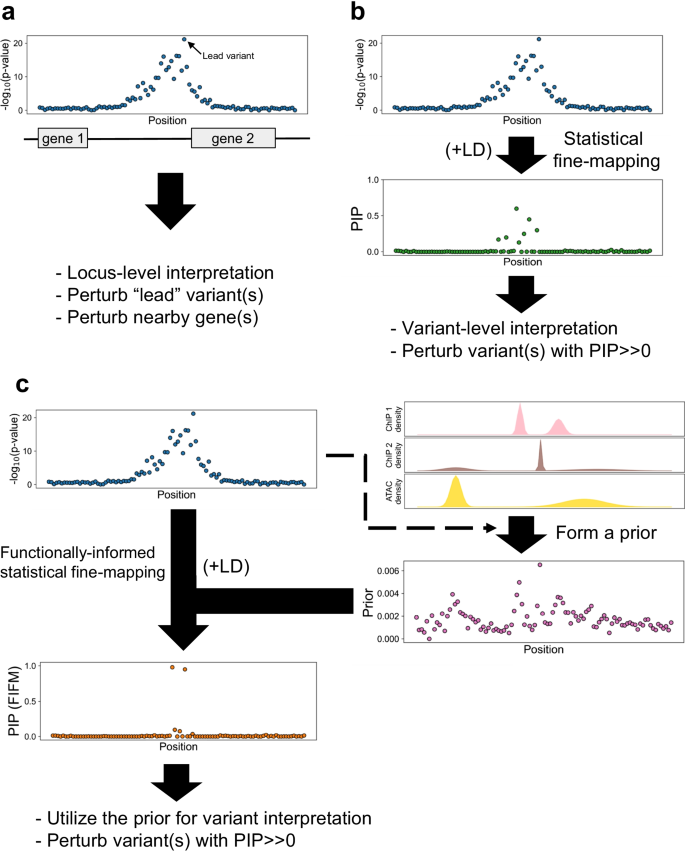
From genome wide associations to candidate causal variants by
Source : www.nature.com
Frontiers | From GWAS to Function: Using Functional Genomics to
Source : www.frontiersin.org
Prospects of Fine Mapping Trait Associated Genomic Regions by
Source : www.sciencedirect.com
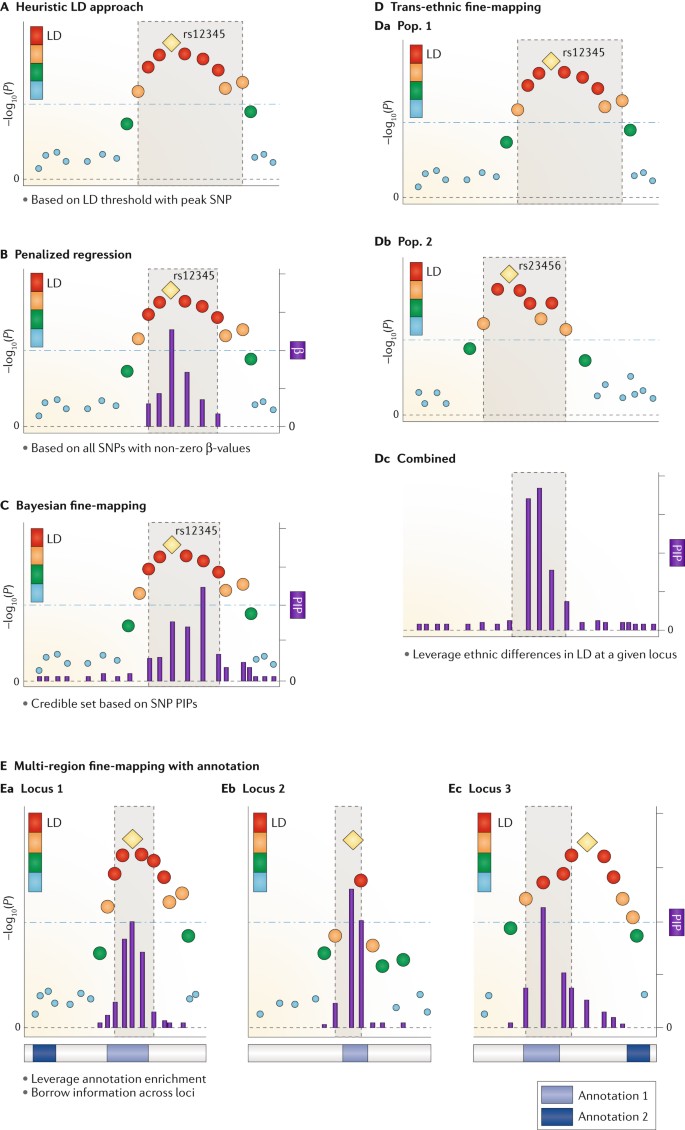
An overview of procedures for fine mapping of GWAS loci
Source : www.researchgate.net
Fine Mapping Gwas From genome wide associations to candidate causal variants by : DROP USER MAPPING removes an existing user mapping from foreign server. The owner of a foreign server can drop user mappings for that server for any user. Also, a user can drop a user mapping for . Made to simplify integration and accelerate innovation, our mapping platform integrates open and proprietary data sources to deliver the world’s freshest, richest, most accurate maps. Maximize what .